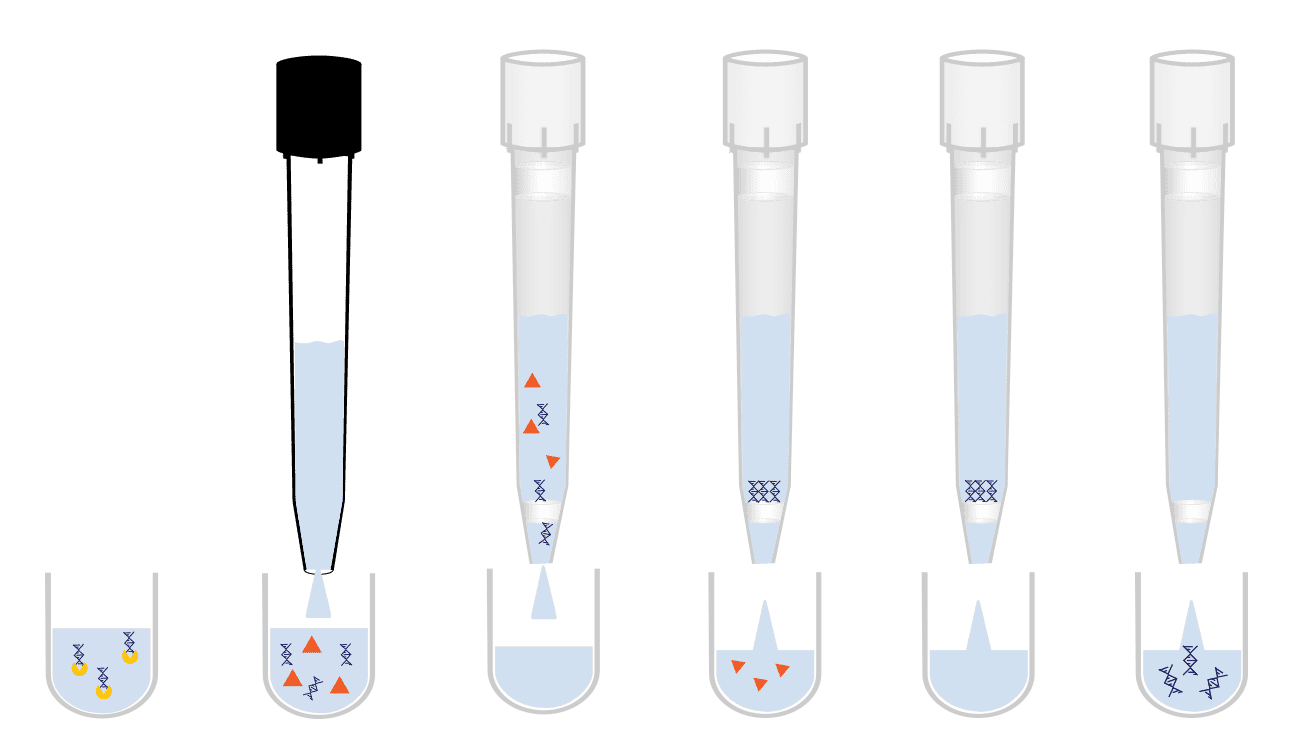
Genomic Sample Preparation
Automated INTip methods for nucleic acid purification from complex biological matrices and cleanup for downstream analysis
How to Order
NiXTips are compatible with major liquid handling robotic platforms. NiXTips are sold in matrix-specific kits for DNA extraction and prep kits for cleanup methods, including necessary buffers. Download product specifications by clicking the applications available. Contact us for custom method development for applications not listed.
NiXTips Applications
Click the application links to download product specification sheets.
★ DNA Isolation from Cell Culture
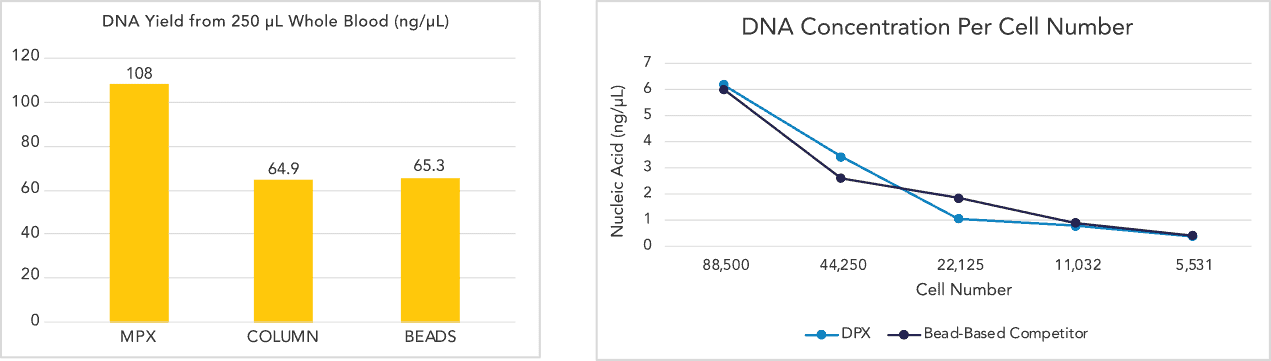
★ DNA Isolation from Whole Blood
★ DNA Isolation from Saliva*
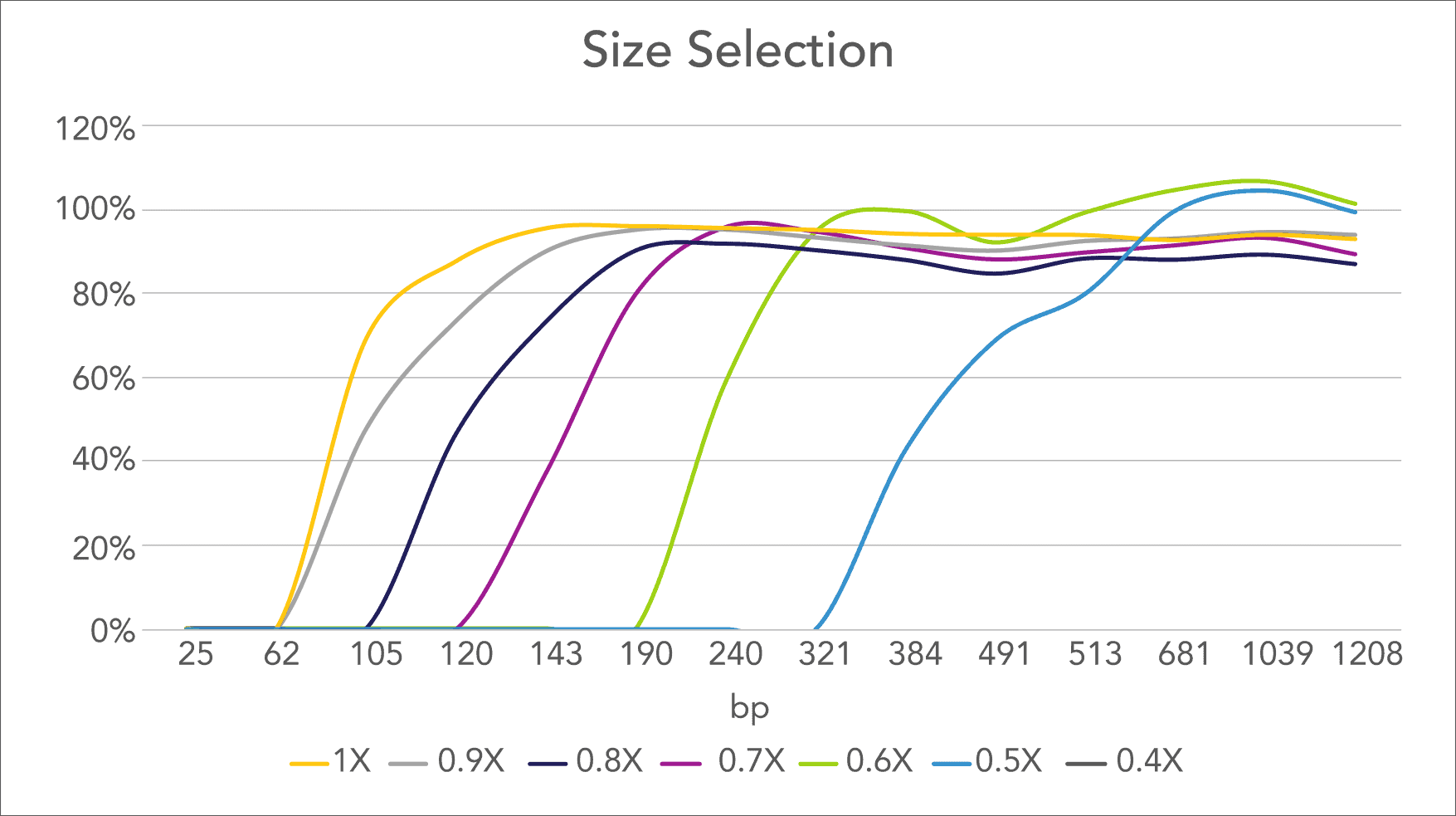
★ PCR Cleanup, Library Prep, Size Selection
*Saliva method coming soon

Hamilton format tips shown here.

| Tip Size -96 tips/rack | Application |
|---|---|
| 50 µL | ★ |
| 300 µL | ★ ★ ★ |
| 1 mL | ★ ★ ★ |

| Tip Size | Application |
|---|---|
| 125 µL -384 tips/box | ★ ★ ★ |
| 300 µL -96 tips/rack | ★ ★ ★ |
| 1250 µL -96 tips/rack | ★ ★ ★ |

| Tip Size -384 tips/box | Application |
|---|---|
| 70 µL | ★ |

| Tip Size -96 tips/rack | Application |
|---|---|
| 200 µL | ★ ★ ★ |
Tecan style NiXTips are coming soon!